Development & code tracking on Ohloh
Posted: August 10, 2012 Filed under: Uncategorized | Tags: ohloh Leave a commentOhloh is another cool site we’ve been using to track the development of PhyloSift. You can visit our Ohloh project page at https://www.ohloh.net/p/PhyloSift (also accessible via the quicklinks sidebar on the right side of this page). Here are some interesting tidbits you can find there (stats accurate as of 8/8/12):
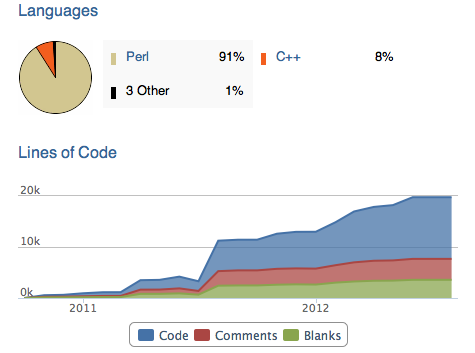
A visual breakdown of the PhyloSift code and its expansion over the last year:
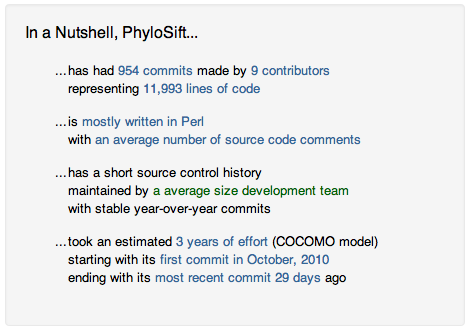
Some interesting statistics:
And Ohloh’s estimate of the overall project cost:
Daily PhyloSift Benchmarks
Posted: July 31, 2012 Filed under: Uncategorized Leave a commentAs we code away here at UC Davis, we’ve been keeping track of PhyloSift’s accuracy and performance using daily benchmarks on simulated data. These benchmarks are carried out using the latest versions of the PhyloSift code available on github (master and development branches).
Click the link below to access graphs showcasing a history of how the PhyloSift pipeline has changed during the course of its development. Here we provide information for simulated Illumina and 454 data, as well as information about PhyloSift compute time for each dataset (running on a single-core).
Daily PhyloSift benchmarks on Simulated Data
Note that these benchmarking plots are permanently accessible via the “Quick Links” sidebar located on the righthand side of the PhyloSift homepage.
Support for Multisample Comparisons
Posted: June 14, 2012 Filed under: Uncategorized Leave a commentMost researchers use barcoded adaptors to pool DNA/RNA from multiple samples and sequence them in a single run (e.g. multiplexing 20 samples within one lane of an Illumina flow cell). We are working hard to increase support for multiplexed data within PhyloSift. Currently, the main client workflow (PhyloSift all mode as executed via the wrapper script) assumes that an input file contains sequence data from a single sample.
We have now prepared a guide to help users conduct multisample comparisons in PhyloSift:
Processing Multiplexed Samples in PhyloSift
Once raw data are demultiplexed and sequence data from individual samples are run through the PhyloSift workflow, the guppy software packages enables rapid taxonomic comparisons across multiple samples. Diversity analyses in guppy include: Principal Components Analysis (akin to PCA using UniFrac), Squash Clustering (akin to UPGMA clustering using UniFrac) and Kantorovich-Rubinstein distance (akin to weighed UniFrac).
New! Workflow Diagrams and Tutorials
Posted: June 4, 2012 Filed under: Uncategorized Leave a commentWe’ve been working hard to improve PhyloSift these past few months, and things are really starting to heat up! You’ll notice some heavy website updates to go accompany our expanded and streamlined code. The following links are permanently housed under Tutorials–>Running Phylosift
For biologists wanting to run their own environmental metagenome data through PhyloSift, we’re compiling a quickstart guide and step-by-step tutorial using test data:
Running Phylosift – An Overview
Metagenome Analysis Tutorial using PE-Illumina Data
PhyloSift also includes discrete workflows for users to incorporate their own marker genes, mine genome data, and generate simulation datasets. Visual diagrams explaining these PhyloSift features can be found here:
Building PhyloSift marker packages (The Monkey)
Generating simulated datasets (The Kangaroo)
Expanding PhyloSift marker databases with new genome data (UpdateDB)
User Support Forum on Google Groups
Posted: March 9, 2012 Filed under: Uncategorized Leave a commentAs we’ve been ramping up the coding, we’re trying to expand our online presence to best meet the needs of our users. With that in mind, we’ve established a dedicated support forum on Google Groups to interact with users and build an online community:
https://groups.google.com/d/forum/phylosift
Bigger. Better. Bio-tastic!
Posted: February 16, 2012 Filed under: Uncategorized | Tags: github, twitter Leave a commentWelcome to the new blog of the PhyloSift pipeline, an integrated bioinformatic toolkit for all of your metagenome data analysis needs!
PhyloSift is being actively developed by Aaron Darling, Guillaume Jospin and Holly Bik in the Eisen Lab at UC Davis.
Follow the latest updates on GitHub: https://github.com/gjospin/phylosift
Or, follow us @PhyloSift on Twitter!