Markers Database download change
Posted: January 3, 2018 Filed under: Uncategorized Leave a commentThe markers database has moved to a new location.
This will NOT get automatically downloaded. The automatic download will not work until we release a code fix.
You can download the markers from the following link for now and extract to the location of your choice. By default, PhyloSift will look in the following directory ~/share/phylosift. This path can be modified in the phylosiftrc file.
https://doi.org/10.6084/m9.figshare.5755404.v4
Sorry for the inconvenience.
New PhyloSift release: v1.0.1
Posted: April 8, 2014 Filed under: Uncategorized Leave a commentAnnouncing a new PhyloSift software release, v1.0.1. All website links have now been updated, or you can download this release directly from the following address:
http://edhar.genomecenter.ucdavis.edu/~koadman/phylosift/releases/phylosift_v1.0.1.tar.bz2
PhyloSift paper selected for the PeerJ Picks 2014 Collection
Posted: March 31, 2014 Filed under: Uncategorized Leave a commentWe’re honored that the software paper describing the PhyloSift pipeline has been selected for inclusion in the PeerJ Picks 2014 Collection:
This Collection celebrates the one-year anniversary of PeerJ publishing their first articles. Staff combed through the 500 articles published so far in both PeerJ and PeerJ PrePrints, and picked out 20 personal highlights which represent the scope and diversity of the articles we have published to date. Some articles have over 25,000 views; some already have a significant number of citations; some represent major new findings; some are important papers in open data and open science; some of the PrePrints have multiple revisions; and some are just plain fun! (description from PeerJ’s website)
Click here to browse all the articles selected as 2014 PeerJ Picks
PhyloSift paper now published in PeerJ
Posted: January 9, 2014 Filed under: Uncategorized Leave a commentWe’re delighted to announce the first paper describing the PhyloSift pipeline, published today in PeerJ. In addition to the paper itself, the manuscript reviews are also public (you can read them here – many thanks to Nick Loman and our anonymous reviewers for their help and insight):
Darling AE, Jospin G, Lowe E, Matsen FA IV, Bik HM et al. (2014) PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243 http://dx.doi.org/10.7717/peerj.243
Please read, discuss, and enjoy!
PhyloSift v1.0.0_02 released
Posted: October 18, 2013 Filed under: Uncategorized Leave a commentHot off the presses! We’ve just released Phylosift v1.0.0_02, our newest stable software release. All website links have been updated accordingly, and you can download this release from the following address:
http://edhar.genomecenter.ucdavis.edu/~koadman/phylosift/releases/phylosift_v1.0.0_02.tar.bz2
This new release includes some major code updates (see our GitHub Issues page for detailed information about updates and bug fixes).
PhyloSift Poster at #asm2013
Posted: May 24, 2013 Filed under: Uncategorized | Tags: conference, poster Leave a commentThanks to everyone who stopped by the PhyloSift poster at the 2013 Annual Meeting of the American Society for Microbiology (held May 17-21 in Denver, Colorado)!
In case you missed it, our poster is now available on Figshare: “PhyloSift: phylogenetic analysis of metagenomes and genomes”
Announcing PhyloSift v1.0.0 Beta Release!
Posted: November 26, 2012 Filed under: Uncategorized Leave a commentAfter months of hard work, we’re extremely happy to announce the first stable beta release of PhyloSift – version 1.0.0. Click here to download the .tar.gz packaged archive (and note that this link is always accessible on the right hand side of each page in the Quick Links sidebar).
We’ve also overhauled the website in preparation for the v1.0.0 release – take a look at our extensive documentation of command line options, expanded list of file outputs, and updated introductory PhyloSift tutorial.
So what are you waiting for? Go ahead and start analyzing that data!
We’re always interested to hear feedback and comments from PhyloSift users — the easiest way to reach us is @PhyloSift on Twitter). And if you happen to find any bugs, please report them on our GitHub issue tracker.
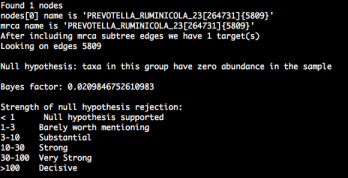
Bayes factor test for taxon presence/absence
Posted: November 13, 2012 Filed under: Uncategorized Leave a commentWe’ve updated PhyloSift to include a new mode: test_lineage. This allows users to conduct a Bayes factor test, a statistical method to test for the presence/absence of a specific taxon amongst the input data (e.g. whether a species might be present in an environmental metagenome). Because the summary files report probability masses for sequences over a reference guide tree topology, not every reported taxon represents a true biological signal (for example, branches where placed sequences have very low probability scores). In conjunction with guppy tree visualizations, the Bayes factor test will help users determine if a specific taxon of interest is likely to be present in their metagenome sample.
Further details on Bayes factor tests can be found on our new tutorial page, and our guide to running PhyloSift on the command line.
Dynamic quality trimming for raw FASTQ input data
Posted: October 8, 2012 Filed under: Uncategorized Leave a commentExciting news! PhyloSift now supports dynamic quality trimming of raw FASTQ data. Upon detecting FASTQ input files, the software workflow will perform Heng Li’s BWA quality trim algorithm (when running PhyloSift all or search mode). Reads are trimmed according to the following formula (where l is the original read length):
argmax_x{\sum_{i=x+1}^l(INT-q_i)} if q_l<INT
To utilize this new feature, download the development version of PhyloSift, available at http://edhar.genomecenter.ucdavis.edu/~koadman/phylosift/devel/phylosift_latest.tar.bz2
Advanced user settings – PhyloSift Run Control file
Posted: August 14, 2012 Filed under: Uncategorized Leave a commentAdvanced users may want to alter the default parameters and search values executed in the PhyloSift pipeline. We’ve added support for a PhyloSift Run Control input file (the phylosiftrc file, which comes packed in the home directory of the PhyloSift download). This file is for advanced users wanting to change the specific settings for different programs packaged within PhyloSift (HMMer, LAST, etc), or for system administrators overseeing a single shared copy of PhyloSift and its reference databases. To use the RC file, specify the command line flag –config=<file>
Further details can be found here: